
I would particularly like to thank my family and friends: especially Natalia and Arno for supporting and encouraging me when I was at home, and even more when I was away from home. Thanks to my advisor Rashid Ibragimov, for his continued support even after leaving University, and for convincing me to expand upon my bachelor thesis. Jan Baumbach for giving me the topic and supervising this thesis even though his group at Saarland University no longer exists.
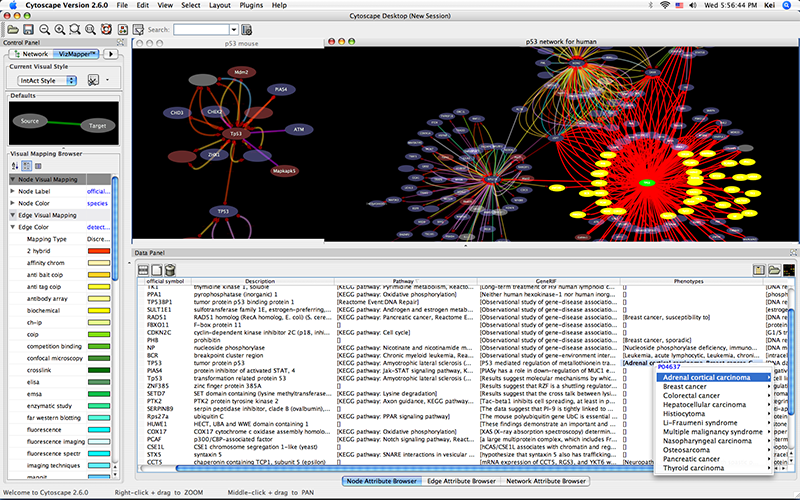
Human, yeast, and fruit fly PPI networks were aligned using GEDEVO, MI-GRAAL, and NETAL, and the results analyzed with CytoGEDEVO, focusing on biological significance.Īcknowledgements I would like to thank everyone who supported me in the course of writing this thesis. Besides the GED, GEDEVO offers additional topological optimization criteria and can also incorparate external data, such as scores derived from Gene Ontology (GO) terms or protein sequence alignments. GEDEVO uses an Evolutionary algorithm (EA) to optimize an alignment topologically by heuristically minimizing the induced Graph Edit Distance (GED). For this purpose, the original GEDEVO aligner has been extended to CytoGEDEVO, a Cytoscape 3 plugin, and enhanced with additional functionality. The main work of this thesis is a visual alignment approach, that involves the user in the alignment process, and provides an easy and intuitive way to perform and visualize alignments. In the last few years many algorithms and tools have been presented to solve this task, but none of them is particularly user friendly, and cannot be used to discover new insights in an intuitive way. This thesis focuses on the global network alignment problem for pairs of PPI networks: Given two input networks, find the best corresponding partner in the other network for each protein as defined by some criterion. However, analysis of this data still poses a problem, as many methods for analyzing networks and gaining insight require solving NP-hard problems. Saarbrücken, 27th January 2015Ībstract Recent years have seen a huge amount of biological network data becoming available, including Protein-Protein-Interaction (PPI) networks, metabolic networks, and gene regulatory networks. Statement I hereby confirm that this thesis is my own work and that I have documented all sources used. At each step of data analysis, Orthoscape also provides for convenient visualization and data manipulation.Ĭytoscape plugin Evolution Gene regulatory network Metabolic pathway Ortholog Paralog Phylostratigraphy.Saarland University Center for Bioinformatics Master’s Program in BioinformaticsĬytoGEDEVO: A Cytoscape app for fast and interactive network alignment Orthoscape allows its users to analyze gene networks or separated gene sets in the context of evolution. The distinctive feature of Orthoscape is the ability to control all data analysis steps via user-friendly interface. Orthoscape aids in analysis of evolutionary information available for gene sets and networks by highlighting: (1) the orthology relationships between genes (2) the evolutionary origin of gene network components (3) the evolutionary pressure mode (diversifying or stabilizing, negative or positive selection) of orthologous groups in general and/or branch-oriented mode. We have developed a new Cytoscape 3.0 application Orthoscape aimed to facilitate evolutionary analysis of gene networks and visualize the results. Nevertheless, only three plugins tagged 'network evolution' found in Cytoscape official app store and in literature. Among them, Cytoscape ( ) is one of the most comprehensive packages, with many plugins and applications which extends its functionality by providing analysis of protein-protein interaction, gene regulatory and gene co-expression networks, metabolic, signaling, neural as well as ecological-type networks including food webs, communities networks etc.
#ORTHOLOG LAYOUT IN CYTOSCAPE SOFTWARE#
There are many available software tools for visualization and analysis of biological networks.


 0 kommentar(er)
0 kommentar(er)
